Eleventh Annual Arkansas Space Grant Symposium, April 25, 2003, Conway, AR. Abramova, T., D.F.Gilmore, K. Halcom, J. Greeno, B. Hogue, and H. Davis, "Studies on the Bacterial Degradation of the Detergent Igepon"
Dr. David F. Gilmore
Environmental Microbiology Research Lab
Bacterial Ecology of Animal Systems
Research on Biodegradable Plastics
General Microbiology
Sulfur Metabolism of Bacteria
Research is always a valuable experience for an undergraduate student (and a necessary one for a graduate student). Not only does it look good on a resume, but the experience itself is very valuable, and it provides hands-on experience doing "real" science that is less structured, but more challenging and informative than a laboratory course. Research connects the information and theory learned in the classroom to real life science.
I have several research projects going on any at one time; some are related,
and some are not. There is always something for an undergraduate to do,
and there are parts of the research that can be tailored into specific projects
for undergraduate Honors Thesis or for graduate study. Potential Masters
degree students are welcome. The ASU Department of Biological Sciences is a very
active department and provides opportunities for research that connect
microbiology with other areas of biology. Please contact me if you would like further
information.
dgilmore@astate.edu
870-972-3263
Bacterial Ecology of Animal Systems
Animal = Humans
We are in the midst of a project studying the prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) among Nursing and Allied Health students. MRSA has been all over the news lately, as community-acquired staph infection resistant to treatment with beta-lactam antibiotics (CA-MRSA) are becoming common. We have over 50 isolates of S. aureus currently being screened for antibiotic resistance. Read more.
Animal = Houseflies
Houseflies and other "filth flies" are recognized as transmitters of infectious disease. We are working with Tanja McKay to experimentally infect houseflies with Escherichia coli and determine how
Biodegradable Plastics Research
Various bacteria synthesize and store a kind of biodegradable plastic, poly(3-hydroxyalkanoate). Much of my research has involved this material. I have written a document about the chemistry, biology, and history of biodegradable plastics to acquaint students with the field. An excerpt is posted here.
Previous research on biodegradation of plastic:
Lee, Kwang-Min, D. F. Gilmore, and M.J. Huss. 2005.
Fungal Degradation of the Bioplastic PHB (Poly-3-hydroxybutyric acid). J.
Polym. Environ. 13(3):213-219.
Timmins, M..R., D. F. Gilmore, N. Lotti, M. Scandola, R.C. Fuller, and R.W. Lenz. 1997. A Spectrophotometric method for detection of enzymatic degradation of thin polymer films. J. Environ. Polym. Degrad. 5(1):1-16.
Gilmore, D.F., N. Lotti, B. Schneider, M. Scandola, R.W. Lenz, and R.C. Fuller. 1994. Biodegradability of blends of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) with cellulose acetate esters in activated sludge. J. Environ. Polym. Degrad. 2:49-57
Gilmore, D.F., S. Antoun, R.W. Lenz, and R.C. Fuller. 1993. Degradation of poly(beta-hydroxyalkanoates) and polyolefin blends in a municipal wastewater treatment facility. J. Environ. Polym. Degrad. 1:269-274
Space Grant Research: Degradation of the Detergent Geropon (formerly "Igepon") and Synthesis of Biodegradable Plastic (PHA)
Supported by grants from the Arkansas Space Grant Consortium
For a space station or extraterrestrial colony to be viable, there are a number of critical life support systems that must be in place, and among them is a system for recycling water. Water is essential for human health, and because of the exorbitant cost of supplying such a colony with any material, water must be recycled on the premises. Much of the wastewater that would be generated is referred to as gray water and is primarily personal hygiene water generated from showering, laundry, etc. As such, the major organic component of the water would be whatever detergent is used for cleaning. Scientists working for NASA developed the detergent Igepon (since, sold to another company and renamed Geropon) although it is not clear whether this detergent will ultimately be used. There are a variety of methods for removing such contaminants from the water including the use of mixed microbial cultures to carry out the complete degradation of the detergent molecule.
We propose an alternative approach that could be used in conjunction with other methods, to find a bacterium that can utilize the detergent as sole carbon source and convert the carbon to biodegradable plastic. In this way not only could the water be partially purified, but the detergent could be converted into a potentially useful product. Items made from this microbially-produced plastic (PHB), once their functional use was ended, could be composted and the nutrients recycled. This study continues to produce interesting results, many of them representing side-trips off the original research idea.
Isolation and Screening of Igepon-utilizing Bacteria:
2001 Arkansas Undergraduate Research Conference,
April 20-21, Henderson State University, Arkadelphia, AR. Murphy, B., A.
Stewart, H. Harrell, and D.F. Gilmore, "Isolation and Identification of Igepon-Degrading,
Plastic-Producing Bacteria"
Using enrichment culture techniques, several environmental samples were screened for the presence of Igepon-degrading bacteria including soil, water, compost, plant roots, and sewage sludge. After isolation, these bacteria were tested for their ability to respond to increasing concentrations with increased growth, demonstrating the ability to use Igepon as a carbon source. Out of 34 bacteria, 13 clearly showed an ability to grow using Igepon. Of these 13, five were inhibited at the higher concentrations of Igepon tested.
The isolates were also tested for their ability to produce PHA using the fluorescent stain Nile Blue A. Five isolates tested positive for PHA production, and all 5 were among those capable of degrading Igepon. All except one isolate were Gram negative rods. Using the Biolog identification system, several of the isolates have been identified. Among the genera capable of Igepon degradation are species of Ralstonia, Pseudomonas, Comomonas, Ochrobactrum, and Stenotrophomonas.
Selection of a "Star Player": Ralstonia sp ASU1
Tenth Annual Arkansas Space Grant Symposium,
April 26, 2002, Jonesboro, AR. Davis, H., B. Murphy, and D.F.Gilmore,
"PHB Production by a Detergent-degrading Bacterium"
Isolates obtained by enrichment culture with Igepon as sole carbon source were further tested for growth at different Igepon concentrations to ensure their ability to degrade Igepon. One isolate, ASU1, showed a nearly six-fold increase in turbidity, indicating good growth, as the Igepon concentration was increased. An upper growth limit was reached at less than 1% Igepon (w/v), likely due to detergent toxicity. Standard bacteriological tests on ASU1 determined that it is a small, motile, aerobic, oxidase positive, Gram negative rod. Identification of this isolate was not possible using the BIOLOG method indicating that the isolate was not present in the database. However, the isolate was very similar to Ralstonia paucula and yet did not match several other Alcaligenes and Ralstonia species, indicating that it is a previously unknown Ralstonia species.
Ralstonia sp. ASU1 was grown under nitrogen limiting conditions to stimulate possible PHB production. Cells were positive for PHB using the Nile Blue A fluorescent assay, and HPLC analysis showed a PHB content of 167 Fg/mg after two days on N-deficient solid medium. To further study PHB production by this organism, batch cultures were grown for 3 days, and samples were analyzed daily for PHB content. Samples of total biomass and PHB samples were dried and analyzed gravimetrically. The total yield of PHB hovered at roughly 60% of total dry weight during each trial. These results suggest that Ralstonia sp. ASU1 has potential for use in PHB production.
Genetics of Igepon-utilizing Bacteria:
Nordeen, R., T. Mon, and D. Gilmore. 2001. Plasmid
Analysis of Bacteria that Metabolize the Detergent Igepon. J. Ark. Acad. Sci.
55: 185-187.
The 13 isolates shown to use Igepon as a carbon source were screened for
plasmids that might contain the genes necessary for Igepon utilization. Dr.
Russell Nordeen and his lab discovered a few plasmid-containing isolates, but
nothing consistent with Igepon utilization. Further research sought to find
mutants unable to utilize Igepon in an attempt to identify and clone the genes
responsible:
In order to provide a broader understanding of the
metabolism of Igepon, studies of genes involved in Igepon degradation are being
conducted using transposon mutagenesis. Transposons are mobile genetic elements
that can insert into or near genes thus altering their expression. The
transposon Tn5, which encodes resistance to the antibiotic kanamycin was loaded
on the narrow host range plasmid R91-5 to create the suicide plasmid pMO1896.
This plasmid was mated into ASU-13, an Igepon metabolizing strain isolated from
sewage sludge. Kanamycin resistant exconjugants were obtained at a frequency of
10-8 per donor cell. These colonies were screened for lack of growth on media
containing Igepon as sole carbon source by replica plating. Of 329 colonies
screened, one mutant has been identified that appears altered in its growth on
Igepon containing media. Efficiency of Tn5 mutagenesis and growth studies on the
putative mutant will be discussed.
Oddly, no mutants were ever found, suggesting that something about the
utilization of Igepon involves vital genes and loss of one of those genes is
lethal.
Studies of Sulfur Metabolism
Eleventh Annual Arkansas Space Grant
Symposium, April 25, 2003, Conway, AR. Abramova, T., D.F.Gilmore, K.
Halcom, J. Greeno, B. Hogue, and H. Davis, "Studies on the Bacterial Degradation
of the Detergent Igepon"
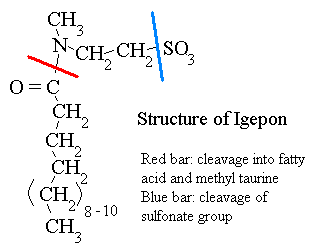
If we are going to study conversion of Igepon to plastic, we must have some understanding of the mechanism of degradation of the molecule. Because the molecule is a sulfonate, and sulfonate degradation is rarely studied (see papers from E.R. Leadbetter, U. Connecticut), we became very interested in the sulfur metabolism of Ralstonia and the other isolates.
The igepon molecule is comprised of fatty acids in amide linkage to N-methyltaurine (NMT) which contains the sulfonate group. Ralstonia ASU1 appeared unable to utilize sulfate or cysteine as S sources, but was able to use methionine and Igepon. Another sulfonate, cysteic acid, was also utilized. Our working hypothesis was that igepon is cleaved at the amide bond, and the S containing product, NMT, is subsequently desulfonated (red line, above). However, neither Ralstonia ASU1 nor several other isolates able to use Igepon as S source were able to grow with NMT. This could be explained in one of two ways. Either our proposed pathway is correct, but NMT cannot be transported into the cell so it is not utilized, or some other degradation pathway is involved.
The possibility of Igepon serving as a N source has been difficult to approach experimentally because the best S sources for Ralstonia ASU1 all happen to contain N (cysteic acid, methionine, Igepon itself).
Analysis of Igepon Usage
Because Igepon is a detergent and potentially toxic to the
bacterium, it is necessary to first determine what concentration is toxic.
Previous experiments showed that Ralstonia ASU1 can make up to 60% of its dry
weight in plastic. More plastic could possibly be produced if the amount of
carbon supplied is greater, but concentrations of Igepon higher than 1% are
toxic to the bacteria. Detergent can be added gradually (fed batch
culture), but a reliable assay is needed to be able to monitor the amount of
detergent in the medium. A common way of determining the amount of anionic
detergent is the wastewater procedure known as the MBAS assay (methylene blue
active substance). This requires large volumes of samples as well as large
volumes of chloroform.
To deal with this problem we miniaturized the MBAS assay and decreased the production of chloroform waste by 20 fold. The standard curve is linear for the range of 0.5 - 10 micrograms of igepon with a correlation coefficient of 0.994.
We were surprised to find that Ralstonia ASU1 was not running out of carbon source in our PHA synthesis experiments, in fact, adding more Igepon only slightly increased the amount removed from the culture medium.
<to be continued>